Click on a chapter to get started.
The use of next-generation sequencing (NGS) for routine clinical care is rapidly accelerating.1,2 This shift is due in part to advances in NGS technology, which have propelled the discovery of somatic mutations that play a pivotal role in hematological disorders and the associated development of targeted therapies.2 These newly identified genetic alterations and molecular pathways provide valuable clinical insights across the continuum of care.2
Hematology has long been the proving ground for molecular testing.1,2 Consequently, the clinical value of next-generation sequencing (NGS) is most apparent today in myeloid molecular testing.1,2
Traditional molecular testing involves iterative single-analyte testing modalities, complex workflows, and frequent outsourcing to referencing laboratories, all of which results in variable, often lengthy, turnaround times.1,3 Associated delays in obtaining results can postpone diagnosis and treatment, negatively impact disease management, and be stressful for patients.3
With an ever-growing list of biomarkers, inherent genetic complexity, and the risk of rapid progression, myeloid malignancies challenge the current iterative testing paradigm and call for a streamlined testing approach that yields rapid results.1
Currently, NGS enables cost-effective, efficient in-house testing of multiple biomarkers on targeted gene panels via streamlined, automated workflows.3 Results can be available within hours or days, depending on the platform.3 With its demonstrated clinical utility in myeloid malignancies, NGS is transforming the testing paradigm and enabling better outcomes for patients.1-3
By leveraging the power of NGS to inform diagnosis, risk stratification, therapy selection, and monitoring in hemato-oncology, progressive pathologists are leading the way in precision medicine. 1,3
Myeloid malignancies arise from mutations in hematopoietic stem or progenitor cells.4 These clonal disorders often exhibit high degrees of heterogeneity, complex karyotypes, and multiple categories of somatic mutations.1,2,4
The natural process of blood cell formation, hematopoietic stem cell differentiation, and generation of myeloid and lymphoid cell lineages.5 Hematopoietic disruptions in the myeloid lineage can lead to 3 major disease categories: acute myeloid leukemia (AML), myeloproliferative neoplasms (MPN), and myelodysplastic syndrome (MDS).4
An aggressive disease (rapid onset and progression) that occurs primarily in adulthood and is marked by an abnormal increase and accumulation of myeloblasts (immature myeloid cells) in the bone marrow and blood, which leads to impaired hematopoiesis and bone marrow failure.6,7
Collectively characterized by an abnormal increase in multiple blood lineages (granulocytes neutrophils, eosinophils, and myelocytes) that exhibit variable degrees of cellular maturity, especially in bone marrow and blood, CMDs also have the potential to undergo clonal evolution.8,9
Myeloproliferative neoplasms present with the clonal proliferation of 1 or more myeloid cell lineages.10 The role of genetic and genomic aberrations in pathogenesis has been well documented for these disorders.1,2 The 4 primary disorders of MPNs are chronic myeloid leukemia (CML), polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF).10
This group of heterogeneous bone marrow disorders is characterized by defective hematopoiesis, growth, and maturation of blood-forming cells, resulting in an abnormal reduction of 1 or more types of blood cells in the bone marrow.11 MDS present with bone marrow failure and associated abnormal cell morphology. 12,13 They also have a high propensity to progress to acute myeloid leukemia (AML).12,13
Characterized by excessive, abnormal white blood cell (granulocyte) production and the presence of the Philadelphia chromosome/BCR-ABL mutation, chronic myeloid leukemia (CML) is a slow-growing cancer of the blood-forming tissue (bone marrow).10,14 The eventual accumulation of immature white blood cells (myeloblasts or blasts) in the blood and bone marrow impairs other blood cell development and leads to a shortage of red blood cells (anemia) and platelets.15
The overproduction of red blood cells characterizes polycythemia vera (PV), 1 of the 3 commonly classical Philadelphia chromosome-negative, or BCR-ABL, myeloproliferative neoplasms.10 PV can result in abnormally thick blood, leading to arterial or venous thrombosis and bleeding.10 About 95% of patients with PV have the Janus kinase 2 (JAK2) V617F.10
Characterized by the overproduction of abnormal blood cells (megakaryocytes) and platelets, essential thrombocythemia (ET) carries an associated increased risk of blood clots and hemorrhagic complications.10 The Janus kinase 2 (JAK2) V617F mutation is found in 50% to 65% of patients with ET.10
Primary myelofibrosis is marked by scar tissue (fibrosis) buildup in the bone marrow, which interferes with normal hematopoiesis, resulting in low blood cell counts.10
About 50% of patients with PMF have the Janus kinase 2 (JAK2) V617F mutation.10
Due to their genetic complexity and high degree of heterogeneity, myeloid malignancies can be challenging for molecular analysis.1 Since some myeloid disorders can progress rapidly, the European LeukemiaNet (ELN) recommends that molecular testing for some biomarkers (eg, NPM1 and FLT3) be available on a rapid-turnaround basis, often within 48 to 72 hours.1,15
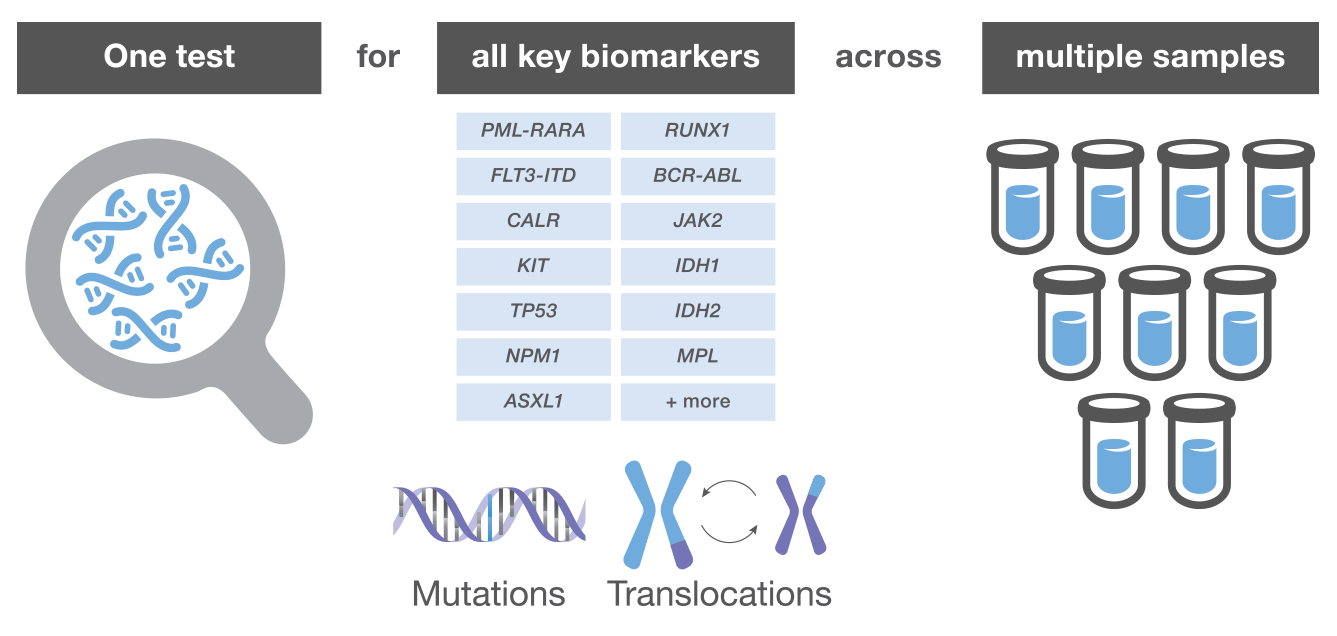
Over the last decade, whole-genome and whole-exome sequencing studies have revealed a number of recurrent mutations associated with these disorders. As the list of biomarkers expands so do the molecular testing algorithms, making it impractical—if not implausible—for laboratories to keep pace using single-gene testing methods.1,16 NGS helps overcome these challenges by empowering labs to perform a multi-gene assessment with a single test and enabling rapid results.1,16
Ultimately, real progress depends on the ability of all molecular testing labs (institutional, hospital, and community) to keep pace and to deliver the answers that clinicians need at each stage of the care continuum. Realizing the promise of precision oncology and its potential for better outcomes in hemato-oncology and beyond entails a new molecular testing paradigm—one where in-house, streamlined biomarker profiling with rapid results is not only possible, but ubiquitous.
Next-generation sequencing
for molecular hematology testing overview.
Megan Lim, MD, PhD - Director of Hematopathology
University of Pennsylvania
The complexities of hemato-oncology testing are evident in AML.1,15 This traditional diagnostic scenario depicts a representative process of testing multiple biomarkers across several modalities with associated staggered turnaround times (TATs), all of which can contribute to deleterious delays in therapy selection and initiation.1

From a clinician’s perspective, the biggest challenges of serial single-gene testing and NGS outsourcing are the variable turnaround times and associated delays in patient care.16,17 A complete mutational profile can take weeks to deliver. For acute and aggressive hematological malignancies, clinicians often require STAT (rapid turnaround) testing to enable timely identification of disease subtype and treatment plan development.1,15

From the lab’s perspective, the limited scope of single-gene assays can be a significant drawback as each test requires time, resources, and separate samples. Additional disadvantages of Sanger sequencing, for example, are its low sensitivity and inability to detect subpopulations reliably or work efficiently with limited sample quantities.16 As such, sequential single-gene testing and NGS outsourcing can take a considerable toll on a lab’s labor and overhead costs.16
From an assay development perspective, aside from being highly resource-intensive, it’s not feasible that laboratories can keep up with the rapid pace of biomarker discovery.16
Consequently, the complexities, costs (tangible and intangible), and impracticalities of performing and developing multiple single-gene assays are beginning to undermine their benefits.16
In-house NGS testing overcomes these challenges by enabling labs to profile all relevant genetic aberrations simultaneously in a massively parallel fashion. NGS generally provides higher sensitivity, larger scale, and the ability to detect novel aberrations compared to traditional methods (Table 1).18
In fact, numerous commercially available panels feature target probes for all key mutations. From 1 test, a lab can identify all relevant point mutations, insertions, deletions, translocations, and copy number variations associated with a given disorder.16 There are panels for DNA targets and RNA targets, which are necessary to detect gene fusions.16 Some platforms even allow simultaneous assessment of DNA and RNA. The primary clinical benefit of in-house NGS testing is that labs can produce a complete mutational profile in as fast as 1 day with the latest technologies.
| Test | Application | Benefits | Limitations |
| Sanger Sequencing | Interrogation of a single gene – must have a known gene region of interest | • Cost effective • Quick and simple workflow |
• Sensitivity (only down to ~20% allele fraction) • Low discovery power • Not cost-efficient for large regions of DNA • Low scalability |
| qPCR | Analysis of specific gene variants at specific locations – researcher must know exactly what and where to interrogate | • High sensitivity • Quick and simple workflow • Easily acquired equipment |
• Limited to only a few gene variants or fusions • No discovery power |
| Targeted NGS | Multiple gene variants across targeted areas of the genome (tens to hundreds of genes) | • High sensitivity • High discovery power (comprehensive genomic coverage) • Greater variant resolution • More data from smaller amounts of DNA • Higher throughput |
• Requires data-handling workflow or bioinformatics • Smaller number of samples may be less cost-effective |
Takeaway: Sanger sequencing and qPCR are best leveraged to interrogate small regions of DNA.18 To screen multiple samples and detect variant types across specific areas of the genome, targeted NGS is more efficient and cost-effective.16

Dr. Bekim Sadokovic
Western University
Quote taken from:
Eur J Haematol. 2019 Sep;103(3):178-189.
The classic approach to molecular hematology testing involves targeted single-analyte testing after initial pre-screening by other diagnostic modalities including flow cytometry, anatomic pathology, and clinical cytogenetics. This approach presents several limitations:
"Given that whole genome or exome sequencing has revealed a wide but limited-range of high-frequency mutations associated with hematologic malignancies such as myeloid neoplasms, including DNA sequence mutations and gene fusions, a DNA/RNA-based NGS panel provides an ideal alternative approach for comprehensive testing of patients as an initial, tier-1 screen in place of targeted triage, rather than as a secondary (tier-2) assay reflexive to other tests."

WGS identifies every DNA base across the entire genome (coding, noncoding, mtDNA) and detects all variant types (SNVs, Indels, SVs, CNVs).19 With its massive data set, WGS entails substantial costs, extended time to results (weeks), and complex data interpretation, making it a method better suited for research and discovery rather than routine clinical testing.3,19

Targeting the protein-coding regions of the genome, WES detects coding variants (SNVs, indels).19 Due to the cost, length of time to results (weeks), and the complexity of data interpretation, WES is better suited for research and discovery than routine clinical testing.3,19

WTS identifies genome-wide differential RNA expression (coding and noncoding). By evaluating altered genetic variants and the continuously changing cellular transcriptome, WTS provides valuable information about the cells and transcriptional networks.20 It is commonly used for discovery and gene-expression analysis.20

Targeted-sequencing panels profile a select set of genes that are curated for a specific application or disease.20 Featuring actionable biomarkers and a smaller data set, these panels are commonly used in clinical applications to help inform the diagnosis, treatment, and monitoring of individual patient disease states.3
When it comes to acute hematological malignancies, early diagnosis and personalized disease management present substantial challenges, as does the reliable and timely detection of measurable residual disease (MRD) —essential to guiding therapy choices, monitoring treatment response, and detecting relapse.3 Genomic biomarkers are the key to unlocking this valuable clinical information.1,3,15
While the value of WGS and WES in studying hematological malignancies is recognized, applying these methods for routine clinical testing is impractical due to costs, long turnaround time to results (weeks), and the complexities of data storage, analysis, and interpretation.3
With their actionable biomarkers, quick turnaround times, and lower cost compared to WES, targeted NGS gene panels are best suited for routine clinical applications and especially ideal for identifying mutations and translocations in specific genes.1,3,16 Some advanced NGS platforms deliver robust integrated analysis and reporting tools, empowering clinicians to facilitate patient selection and clinical trial inclusion.12,21
Relying on the high sensitivity of NGS, clinicians can utilize it to monitor multiple mutations over time to better track a patient’s evolving mutational landscape.9,15 NGS promises to advance precision medicine by identifying actionable biomarkers and individual disease mutations and matching patients to clinical trials, all ultimately contributing to the development of novel, targeted therapies, tailored disease management, and better outcomes.1,3,12,21

Given the substantial phenotypic and genetic heterogeneity of myeloid disorders and the continually increasing number of alterations that need to be analyzed, many more laboratories are integrating NGS into their diagnostic algorithms.1,3,17
By providing a complete analysis of genetic alterations, including single-nucleotide variants, translocations, and small insertions and deletions, NGS myeloid panels can enable enhanced differentiated disease classification, risk stratification, and improved therapeutic decisions.1,3,17
Watch a brief workflow overview video below.
Click the slider arrows below to see workflow.
The NGS workflow is initiated by extracting, purifying, and quantifying the DNA or RNA from a specimen. Next, the genomic regions of interest are enriched to produce a library of amplicons or DNA fragments.22
The 2 primary methods of library preparation are hybrid-capture and amplicon-based enrichment.22,23
With hybrid-capture, as the name implies, complementary DNA probes are used to capture regions of interest.23
In amplicon-based enrichment, PCR primers are used to amplify areas of interest.22
Since amplicon-based library preparation is faster, requires less DNA input, costs less, and can provide better sensitivity for subclonal populations or heterogeneous samples, it has broad utility in hemato-oncology testing.16
NGS enables a lab to prepare a library from a single specimen or to combine (multiplex) specimens to reduce per-sample costs and scale-up as needed. With sample multiplexing, specimens are barcoded for simultaneous sequencing and then individually analyzed during the results stage.22
Each DNA base is sequentially identified in a massively parallel fashion across the entire library. The 2 most common sequencing methods are light-based technology and ion semiconductor.22
The most advanced NGS systems are equipped with automated analysis software that produces variant reports and annotates biomarkers with information about medical guidelines, associated therapies, and ongoing global clinical trials.21,22, 24-26
These integrated and streamlined analysis capabilities simplify reporting and help labs overcome the previous challenges of data interpretation.22
In the past, NGS adoption was limited mainly to academic institutions, specialty cancer centers, and large reference labs.27,28 Due to the complexity of the technology, the high costs, and the need for specialized bioinformatics expertise, NGS testing was primarily outsourced when needs arose in the community hospital setting.27,28
Significant advancements in NGS technology, workflow automation, integrated analysis, and reporting software—in some platforms—have leveled the playing field, making in-house NGS accessible for most community hospitals and labs.22,27
Precision medicine and its promise of improved outcomes will be realized as more and more labs bring the latest highly automated NGS platforms in-house and genetic testing becomes routine in clinical practice.1,27
Hear more from Dr. Morrison on Solving Common Hematopathology Testing Challenges with Next-Generation Sequencing.
Watch 2 minute video clip
Dr. Carl Morrison, MD
SVP, Scientific Development and Integrative Medicine
Roswell Park Comprehensive Cancer Center

Dr. Kojo Elenitoba-Johnson, MD
Peter C. Nowell, M.D. Professor of Pathology and Laboratory Medicine
Perelman School of Medicine at the University of Pennsylvania
Director, Center for Personalized Diagnostics
Director, Division of Precision and Computational Diagnostics
Rapid assessment of genetic biomarkers in myeloid malignancies is vital for early diagnosis, first-line therapy choices, and ongoing clinical disease management.3,29 This includes evaluation of therapy response and relapse/measurable residual disease (MRD) detection.3,29 In particular, acute diseases like AML require rapid test results for key diagnostic and risk stratification markers (ie, PML-RARA gene fusion, FLT3 or IDH1/2, and NPM1).3,15 The 2017 ELN guidelines recommend that results for FLT3 and NPM1 be available within 72 hours.15
"There are several protocols, including extensive amounts of genetics and immunophenotyping, which are required to establish the appropriate approach to treat patients. The sooner we can get results and deliver treatment, the better the outcomes."
Advances in AML risk-stratification guidelines and targeted therapies have made it increasingly important for clinicians to consider a wide range of genetic biomarkers to guide front-line patient-care decisions.3,15,29 NGS helps to address this need by rapidly providing results for a broad number of genetic targets.3 The latest instruments enable a laboratory to sequence a patient sample in about 24 hours so clinicians can have the results in days.3
During a recent UAB study presentation, Craig Mackinnon, MD, PhD, discussed the key considerations for setting up a next-generation sequencing (NGS) panel in a molecular oncology research lab. Basic requirements included the capacity to target genes of interest and detect a wide range of variants, including single nucleotide variants (SNVs), small indels, and fusions, all while accommodating multiple specimen types of varying quality and input amounts.
Dr, Mackinnon concluded that automation, hands-on time, turnaround time, and the associated potential for cost savings should all be part of the evaluation.
Watch 1 minute video clip
Craig Mackinnon, PhD
Director, Genomic Diagnostics and Bioinformatics - Professor, Department of Pathology, University of Alabama at Birmingham
| Consolidates multiple tests into 1 panel | ✓ |
| Detects a wide range of targets | ✓ |
| Accommodates a variety of specimen types (sample quality, input amount) | ✓ |
| Features an automated workflow with minimal hands-on time | ✓ |
| Delivers a rapid turnaround time | ✓ |
| Incorporates automated bioinformatics analysis and reporting | ✓ |
| Includes variant interpretation software | ✓ |
| Offers the potential for cost savings | ✓ |
Advances in NGS have made simultaneous assessment of multiple target genes possible in daily laboratory analysis.1,3 Targeted NGS panels designed explicitly for myeloid neoplasms can inform patient-management decisions across the entire continuum of care—from streamlining diagnosis, identifying disease subtypes, facilitating treatment decisions, assessing risk stratification, and providing prognostic insights to improving monitoring and measurable residual disease (MRD) detection.1,3,17
As in-house NGS testing becomes ubiquitous at a community hospital-lab level, more patients will be able to benefit from these important advancements in precision oncology.

Historically, most community-based labs have utilized traditional single-analyte molecular testing methods, outsourcing to reference labs as needed to augment their in-house capabilities.27,28
This strategy presents a challenge when multiple biomarkers need to be tested, as is the case for myeloproliferative neoplasms (MPNs). Sequential iterative testing can contribute to substantial delays in diagnosis and treatment.1,3,16, 17 These delays can be time and cost-intensive for labs, frustrating for clinicians, and angst-provoking for patients.1,3,16, 17
Today, multi-gene NGS panels enable labs to analyze all relevant genes using a single test that yields results within days.3 With NGS technology delivering expedited results, clinicians will have the information they need to optimize disease management sooner, and patients will have peace of mind.1-3
During a recent conference, Dr. Yi Ding of Geisinger Medical Laboratories described how her laboratory overcame the MPN testing challenge. Before they brought NGS in-house, MPN genetic testing took 4 to 6 weeks in their lab. Patient follow-ups were scheduled as far out as 3 months after the initial visit. By bringing a multi-gene NGS panel in-house, they dramatically reduced the time to diagnosis.
Takeaway: Multi-gene NGS panels that include genes and mutations recommended by National Comprehensive Cancer Network (NCCN) guidelines and deliver timely results offer unprecedented opportunities to streamline diagnosis and enhance disease management for improved outcomes and quality of life.1-3,16,32
Watch 5 minute video clip
Yi Ding, MD, PHD
Systems & Core Laboratory Director of Molecular Diagnostics
Geisinger Medical Laboratories
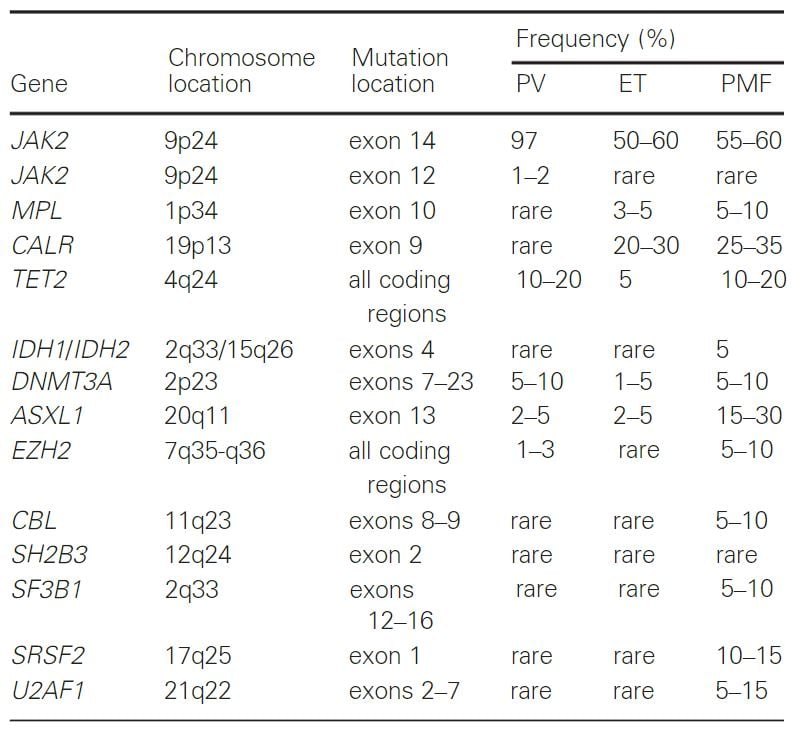
PV, polycythemia vera; ET, essential thrombocythemia; PMF, primarymyelofibrosis
Many myeloid malignancies are classified by the presence of specific genetic mutations.1,3 While an initial diagnosis can be achieved with histopathological and immunophenotyping tests, disease subtypes are usually determined through karyotyping and genetic analysis.1 Per the 2016 World Health Organization (WHO) classification, AML subtypes include recurrent genetic abnormalities and germline predisposition.1,29 In fact, > 95% of AML cases harbor at least 1 somatic alteration.34 In addition, there are approximately a dozen genomic alterations and an average of 3 driver mutations identified per AML sample.34
NGS offers critical advantages in AML disease subclassification as all relevant mutations can be identified in a single test.34 Some of the latest NGS platforms deliver results within hours or days, so these mutational insights can be evaluated in conjunction with the histopathological or immunophenotyping results for a more complete picture of the disease pathology.3
Indeed, the inclusion of certain mutations in the key treatment guidelines (WHO classification, 2016, ELN, 2017, and the National Comprehensive Cancer Network (NCCN)) provides strong evidence for how mutational testing is influencing clinical disease management.18, 29,32,35 Genetic profiling and disease subclassification is expected to become increasingly important in daily clinical practice.31
AML with t(8;21)(q22;q22.1); RUNX1-RUNX1T1
AML with inv (16)(p13.1 q22) or t(16;16)(p13.1;q22);
CBFB-MYH11
Acute promyelocytic leukaemia with PML-RARA
AML with t(9;11)(p21.3;q23.3); KMT2A-MLLT3
AML with t(6;9)(q23;q34.1);DEK-NUP214
AML with inv(3)(q21.3q26.2) or t(3;3)(q21.3q26.2);
GATA2 MECOM
AML (megakaryoblastic) with t(1;22)(q13.3;q13.1);
RBM15-MKL1
AML with BCR-ABL1
AML with mutated NPM-1
AML with biallelic mutation of CEBPA
AML with mutated RUNX1
Therapy-related myeloid neoplasms
AML with minimal differentiation
AML without maturation
AML with maturation
Acute myelomonocytic leukaemia
Acute monoblastic and monocytic leukaemia
Pure erythroid leukaemia
Acute megakaryoblastic leukaemia
Acute basophilic leukaemia
Acute panmyelosis with myelofibrosis
Myelodysplastic syndrome with single lineage dysplasia
Myelodysplastic syndrome with ring sideroblasts and single lineage dysplasia
Myelodysplastic syndrome with ring sideroblasts and multilineage dysplasia
Myelodysplastic syndrome with multilineage dysplasia
Myelodysplastic syndrome with excess blasts
Myelodysplastic syndrome with isolated del(5q)
Myelodysplastic syndrome, unclassifiable
Refractory cytopenia of childhood
Acute myeloid leukaemia with germline CEBPA mutation
Myeloid neoplasms with germline DDX41 mutation
Myeloid neoplasms with germline RUNX1 mutation
Myeloid neoplasms with germline ANKRD26 mutation
Myeloid neoplasms with germline ETV6 mutation
Myeloid neoplasms with germline GATA2 mutation
Transient abnormal myelopoiesis associated with
Down syndrome
Myeloid leukemia associated with Down syndrome
Cutaneous mastocytosis
Indolent systemic mastocytosis
Systemic mastocytosis with an associated haem. neoplasm
Aggressive systemic mastocytosis
Mast cell leukemia
Mast cell sarcoma
Acute undifferentiated leukemias
Mixed-phenotype acute leukaemia with
t(9,22)(q34.1;q11.2); BCR-ABL1
Mixed-phenotype acute leukemia with t(v;11q23.3); KMT2A-rearranged
Mixed-phenotype acute leukemia, B/myeloid, NOS
Mixed-phenotype acute leukemia, T/myeloid, NOS
Mixed-phenotype acute leukaemia, NOS, rare types
Acute leukaemias of ambiguous lineage, NOS
Chronic myeloid leukemia, BCR-ABL1-positive
Chronic neutrophilic leukemia
Polycythemia vera
Primary myelofibrosis
Essential thrombocythemia
Chronic eosinophilic leukemia, NOS
Myeloproliferative neoplasm, unclassifiable
Myeloid/lymphoid neoplasms with PDGFRA rearrangement
Myeloid/lymphoid neoplasms with PDGFRB rearrangement
Myeloid/lymphoid neoplasms with FGFR1 rearrangement
Myeloid/lymphoid neoplasms with PCM1-JAK2
Genetic abnormalities are powerful prognostic factors that can help inform therapy selection.15, 36 For patients with AML, genetic mutations can indicate the suitability for allogenic hematopoietic stem cell transplantation (allo HCT).15, 36 There are also multiple co-occurrent mutations that can impact risk stratification in AML.15 Results from conventional cytogenetics and NGS mutational screenings are routinely used in clinical practice.16
The 2017 ELN guideline groups genetic abnormalities into 3 levels of risk that range from favorable to adverse.15 One of the most common examples of genes with co-occurring mutations that impact risk stratification are NPM1 and FLT3-ITD.15 Risk level is dependent on the presence of both mutations and the allelic ratio of FLT3-ITD.15
2017 European LeukemiaNet (ELN) Guidelines13
| Status | Genetic Abnormality |
| Favorable |
|
| Intermediate |
|
| Adverse |
|
Key Takeaway: Both the NCCN and ELN guidelines recommend that patients with suspected AML be screened for gene mutations.15,32
NGS offers distinct advantages for risk stratification. Given the sheer number of genetic abnormalities that must be evaluated, a multi-gene NGS panel can consolidate many, if not all of the individual single-gene tests that would otherwise be required.3,16 Additional NGS identification of SNVs, insertions, deletions, and rearrangements could provide added value.37
Currently, NGS is the most cost-effective method of risk assessment for AML.3 By utilizing in-house NGS, labs could potentially accelerate time to results while simultaneously lowering costs and reducing the strain on its resources.1,3,16
Detecting leukemic drivers with NGS informs disease management
Over the last decade, NGS has helped unravel the complex molecular underpinnings of myelodysplastic syndromes (MDS).35 Dozens of recurrent mutations associated with the disease have been revealed.35 Targeted sequencing will typically identify at least 1 mutation in the majority of cases.35 While NGS does not replace conventional cytology, histology, and cytogenetics, it can provide meaningful insights from diagnosis and prognosis to treatment response in MDS patients.35
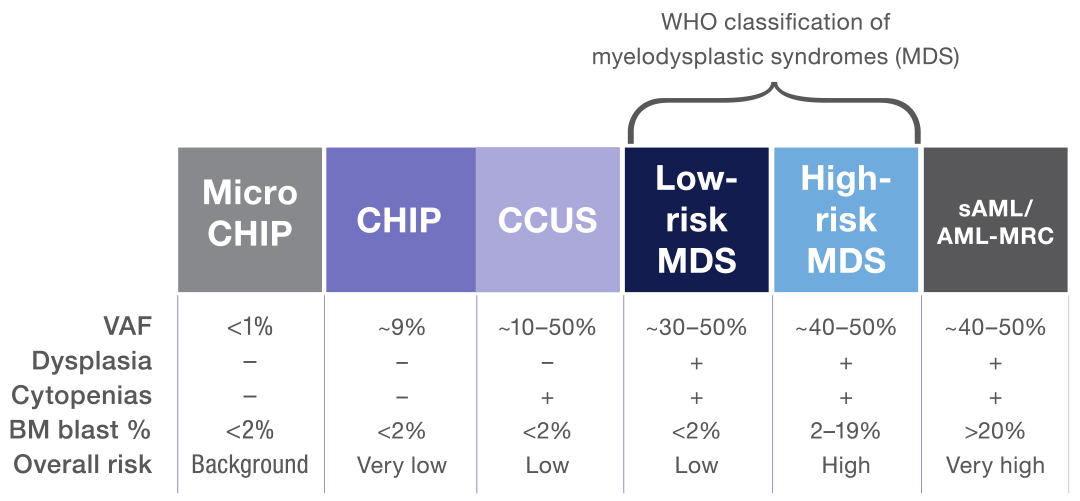
MDS is a clonal disease arising from the proliferation of mutated hematopoietic stem cells.12,35 The spectrum of clonal myeloid disorders ranges from clonal hematopoiesis of indeterminate potential (CHIP) to secondary acute myeloid leukemia (sAML).35,38 MDS is characterized by peripheral blood cytopenia, dysplasia, and the absence of features that define acute leukemia.12,38 In cases where clonal hematopoiesis is detected in the absence of a cytopenia or dysplasia, mutational data can help distinguish MDS from CHIP.38 Typically, a larger number of mutations with a higher variant allelic frequency (VAF) are more supportive of an MDS diagnosis.38
Notably, mutational data of clonal myeloid disorders can also help inform the level of risk for a particular disease to progress to another more serious disorder (eg, clonal cytopenia of undetermined significance [CCUS] evolving to MDS and MDS evolving to sAML).12,38,39
Ongoing research in this area is likely to increase the clinical relevance of NGS for diagnosing and treating these disorders.12,21,35
Since the 1970s, the standard of care for AML has been cytotoxic chemotherapy, and long-term overall survival has remained relatively poor.40 Progress in the development of mutation-targeted therapies in recent years is transforming the AML treatment paradigm.40 Finally, there are more options for patients, and with them, the promise of better clinical outcomes.40
NGS has helped identify many significant genetic alterations, leading to the development of therapeutics that target specific gene mutations.40 In 2017, midostaurin became the first tyrosine kinase inhibitor (TKI) approved for FLT3-mutated AML; many more targeted therapies are emerging, including the IDH2 inhibitor, enasidenib for relapsed/refractory (R/R) AML.40 While further research is needed to understand all the genomic complexities at play, it is apparent that these new targeted therapies are raising the standard of care for AML and improving overall survival and quality of life.40
With its comprehensive genetic profile of the disease, including targetable mutation and other genetic abnormalities, NGS can offer tremendous clinical insights into AML, from risk stratification and therapy selection to disease monitoring.40

Explosion of new AML therapy options in recent years
7+3 induction chemotherapy
AML
Idarubicin
AML
Arsenic trioxide
Target: PML-RARA
APL
Midostaurin
Target: FLT3
Newly diagnosed FLT3-mutated AML
CPX-351
t-AML or AML-MRC
Gemtuzumab ozogamicin
Newly diagnosed and R/R CD33-positive AML
Gilteritinib
Target: FLT3
R/R FLT3-mutated AML
Enasidenib
Target: IDH2
R/R IDH2-mutated AML
Glasdegib
Target: Hedgehog
Newly diagnosed AML (age >75)*
Venetoclax
Target: BCL2
Newly diagnosed AML (age >75)*
Ivosidenib
Target: IDH1
Newly diagnosed IDH1-mutated AML (age >75)*
Oral azacitidine
AML (in first remission)
R/R = Relapse/Refractory
* Patients >75 years old or who have comorbidities that preclude the use of intensive induction chemotherapy
Recurrence monitoring and measurable residual disease (MRD) detection are critical for hematological disorders.1,3,17 While our improved understanding of the molecular landscape of AML has resulted in more informed and targeted first-line treatments, the threat of disease relapse is always looming.40 Studies have shown that negative MRD (assessed by molecular techniques or immunophenotyping) may be a better predictor of survival as it’s associated with a lower risk of relapse.3
Chart: NGS identification of clonal and subclonal mutations in AML provides insights into prognosis, treatment, and response.21
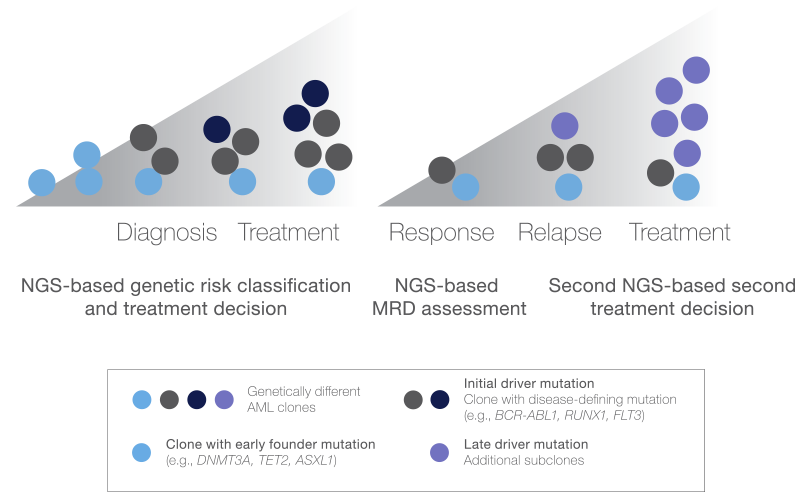
This schematic depicts the subclonal genetic heterogeneity of AML development and progression and illustrates the critical role of NGS in the assessment of clonal and subclonal mutations throughout disease management.
By traditional standards, complete remission is achieved when a morphological assessment shows < 5% myeloblasts (immature myeloid cells) in the bone marrow, absence of circulating blasts, and hematologic recovery.21 Although most AML patients achieve complete remission after primary treatment, many eventually relapse, indicating that morphology is not the best predictor of survival.21
CHART: Studies demonstrate the prognostic value of NGS and multiparameter flow cytometry (MFC) MRD for AML relapse (~75% concordance)41,42 (Source)
Molecular diagnostic techniques, such as multiparameter flow cytometry (MFC) and quantitative real‐time PCR (qPCR) assays, are commonly used to measure and monitor disease burden following treatment of myeloid disorders.41,42
MFC has been the gold standard for MRD detection, but it has drawbacks. For example, it is operator- and center-dependent and without a standardized method of enumerating flow-based MRD in AML.42 Most notably, MFC lacks sensitivity for detecting residual disease.1
While qPCR assays are highly sensitive, most monitoring strategies for AML are limited to identifying the most common AML‐associated mutations and translocations.38 Since only 15% to 25% of AML cases have these known molecular markers, and even fewer MDS cases have them, qPCR offers limited utility.43
Another challenge for qPCR is tracking post-chemotherapy clonal evolution as non-founder clonal mutations may be lost or gained despite the persistence of the founding clones.38 With the considerable time, cost, and resources involved in designing patient-specific primer sets for recurrent somatic single nucleotide variants and small insertions, qPCR is impractical for AML monitoring.1,3,16
Today, more labs rely on NGS‐based tumor-burden monitoring assays because they cover a broader range of genes and have the sensitivity to detect small clonal populations by identifying low‐frequency somatic mutations associated with the malignant clones.1,21,37 Unlike qPCR‐based assays, targeted NGS panels can simultaneously detect many mutated genes and translocations found in myeloid malignancies—without the need to develop patient‐specific primer sets.16,37
Given AML’s combinatorial complexity and mutational combinations (3 to 5 driver mutations in more than 250 genes), NGS offers multiple advantages for MRD.44 NGS can detect the persistence of disease-specific variants to inform treatment and prognosis and identify patients at high risk for early relapse so they can benefit from closer tracking and additional therapies (stem cell transplants).1,3,16, 37,44
Benefits of NGS:

Advances in NGS workflow automation and integrated reporting have empowered clinicians to perform simultaneous assessments of multiple target genes in routine laboratory analysis.1,3,16 An ever-growing list of biomarkers and their inherent genetic complexity have made myeloid malignancies the proving ground for this new testing paradigm.1,2
The value of integrating NGS into daily clinical practice for rapid, streamlined analysis and utility across the care continuum is no longer in question.1,3,15,16,21,29,32 Assessment of myeloid neoplasms with targeted NGS panels is demonstrated to improve diagnosis, assist therapeutic decisions, inform prognosis, and better detect measurable residual disease.1 Adoption of in-house NGS brings advanced molecular diagnosis and precision oncology to more patients and will help raise the standard of care.1-3,12,27,32,40

Historically, cancer treatment was limited to a one-size-fits-all model, where therapies were selected based on the cancer type (site) alone. Today, NGS has advanced the understanding of cancer biology so many therapy decisions can be based on specific cancer biomarkers, and genetic alterations in an individual’s cancer genome.1-3,13,24
Empowered with a molecular profile of a patient’s cancer, clinicians can tailor treatments to optimize outcomes and prescribe therapeutics that directly target biological pathways while avoiding sub-optimal therapies.1-3,13,24
Imagine every oncology patient receiving treatment individualized to their specific cancer genome. What was once a vision for improved cancer care is finally a reality—a world where molecular insights transform clinical care, improve outcomes, and enhance the quality of life for patients.

We've detected your location to be Japan.
Sorry, you cannot access this website. The content on www.oncomine.com is only intended for healthcare professionals. Formore information on our research solutions, please visit ThermoFisher.com
このウェブサイトは、日本国内の医療関係者の方への情報提供を目的としており、一般の方に対する情報提供を目的としたものではないことをご了承ください。研究用製品の情報はThermoFisher.comよりご覧ください。