
Craig Mackinnon, MD, PhD, Director of Genomic Diagnostics and Bioinformatics and Professor of the Department of Pathology, and his team at the University of Alabama, Birmingham (UAB) conducted a study evaluating the performance of the Oncomine Myeloid Research Assay GX on the Genexus System with samples containing mutations in key genes associated with various myeloid research disorders.
Difficult-to-detect targets such as FLT3, CALR, and CEBPA were included. Performance was assessed against the well-established Ion Torrent™ Oncomine™ Myeloid Research Assay on the Ion GeneStudio S5 System.
Dr. Mackinnon advises anyone interested in setting up a next-generation sequencing (NGS) panel in a molecular oncology research lab to consider a platform that can accommodate a wide range of specimens, including those that are small and/or low quantity.
Sequencing challenges are compounded by the diversity of frequently co-existing complex variants underlying somatic changes present in many samples. Consequently, robust NGS assays capable of identifying single-nucleotide variants (SNV), small indels, and fusions in a single workflow are required.
Table 1: A range of commonly mutated genes in myeloid research malignancies.
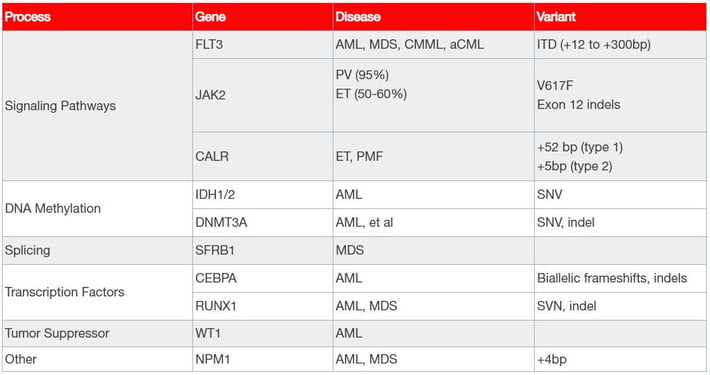
Table 2: Results show high concordance between the Genexus System and the GeneStudio S5 System.
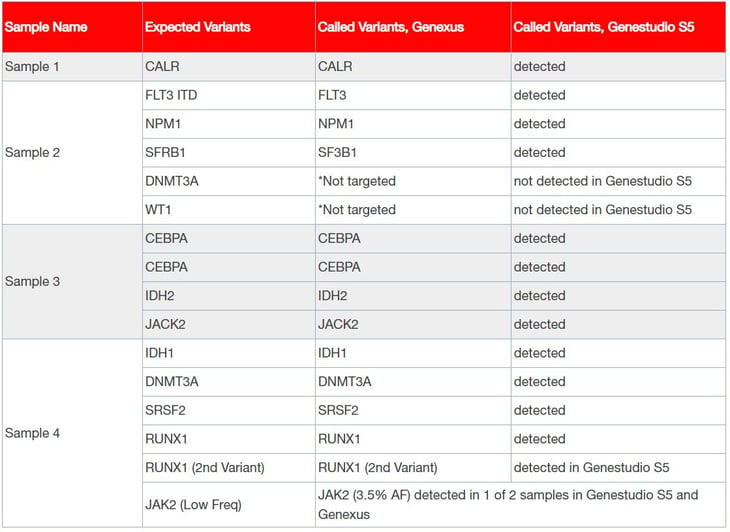
The Oncomine Myeloid Research Assay GX achieved 100% concordance with Myeloid Research Assay on the GeneStudio S5 System
In their quest to optimize their lab operations, UAB evaluated the Oncomine Myeloid Assay GX and the Genexus System. Dr. Mackinnon concluded that the Genexus System and its assays offer several key advantages over previous NGS platforms:
If you would like to learn more about this study, Watch Dr. Mackinnon's Presentation
Precision medicine is rapidly changing our understanding of cancer research and treatment decisions. These breakthrough, personalized treatments hold promise even for patients with historically hard-to-treat diseases, like lung or breast cancer. But
In his recent talk at OncomineWorld 2023, Dr. Bekim Sadikovic presented a strong, evidence-based argument for frontline next-generation sequencing (NGS) in myeloid malignancy testing.
Bekim Sadikovic PhD, DABMGG, FACMGProfessor, Research Chair, and...
In a recent Association for Molecular Pathology (AMP) workshop, Dr. Cecilia Yeung shared how her lab is implementing rapid next-generation sequencing (NGS) to help advance hemato-oncology research and expand access to underserved populations.
...