
Dr. Yi Ding of Geisinger Medical Laboratories recently presented at the 2020 Association of Molecular Pathology annual meeting. During her talk, she described the value of NGS in hematopathology research and shared her experience utilizing the Oncomine Myeloid Research Assay GX and the Genexus System.
As the number of cancer-associated biomarkers has dramatically increased over the last few decades, molecular testing has become a critical component of precision oncology research. In recent years, NGS has played an increasingly important role in hematopathology, owing to its ability to rapidly and simultaneously interrogate multiple genomic aberrations from a single sample.
“Most labs use STAT FISH or RT-PCR for PML-RARA detection because results are needed in <48 hours. Conventional NGS
is not used, as it generally takes 4–5 days for results. With its automated processes and speed—from library preparation to variant interpretation in ~24 hours—the Genexus System has strong potential to create a new testing paradigm.”
Yi Ding, MD, PHD
System and Core Laboratory Director of Molecular Diagnostics
Geisinger Medical Laboratories
Myeloproliferative Neoplasm (MPN) Testing: Single-gene testing vs NGS
Molecular profiling of MPN samples can be particularly challenging due to their underlying genetic complexity. As such, MPN testing is a multifactorial process,
which includes CBC value and molecular testing.
Recently, driver mutations have been identified in more than 90% of MPN samples, which has provided substantial insight into the pathogenesis of these diseases. Testing is generally conducted using the sequential model or with a multigene NGS panel.
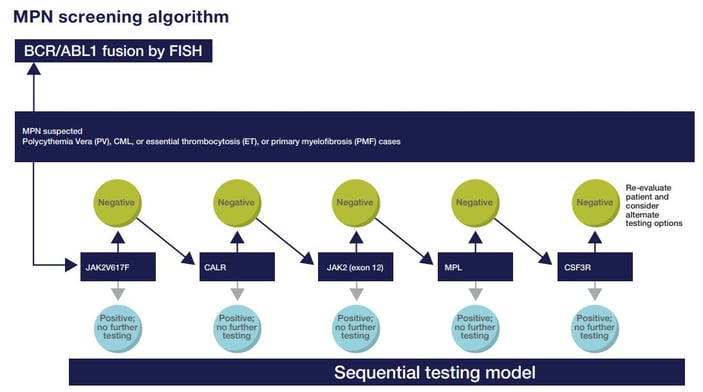
Figure 1. Sequential testing of MPN samples by Geisinger Medical Laboratories. For all samples, testing for the BCR-ABL1 fusion and sequential gene mutation testing was conducted in parallel. JAK2 v617F was tested first. If negative, CALR was tested next and so on. In Dr. Ding's lab, only the JAK2 v617F was conducted in-house so there was usually a 4–6 week turnaround time for results. Since about 75% of the samples are negative, the majority of the patients had to go through the entire testing process, which can drive up costs and extend the time to answers.
Evaluation of the Genexus System for Potential Detection of Key MPN Variants and Translocations
Geisinger Medical Laboratories recently implemented a multigene NGS testing model that includes all the relevant genes and mutations. This approach enabled them to test for all relevant myeloproliferative neoplasm (MPN) mutations in a much more rapid and streamlined manner. As they are always looking to improve their capabilities, the lab recently evaluated the potential of the Oncomine Myeloid Research Assay GX for the detection of key variants and translocations associated with MPNs. With the Genexus System, the assay can yield results as fast as a single day.
Five samples were tested on each platform, including a variety of variants and allele frequencies—current in-house NGS test vs. the Oncomine Myeloid Research Assay GX. They included emerging MPN markers like NRAS and TP53. In addition, they assessed the potential of the assay to detect FLT3-ITD mutations and key fusions for myeloid disorder research.
Oncomine Myeloid Assay GX Evaluation Results:
• Achieved overall sensitivity of 97.1% for key MPN variants
• Demonstrated higher sensitivity than traditional cytogenetic methods for detecting BCR-ABL1 fusions
• Attained accurate detection of common FLT3-ITD mutations
• Delivered good performance for other fusions
Table Below. MPN research: Summary of current in-house NGS vs. the Oncomine Myeloid Research Assay GX on the Genexus System (n = 35).
| Sample | Variant | Current NGS (VAF%) | Genexus Myeloid* VAF%) |
|
GMC-1 |
CALR L367T*46 |
55.3 |
51.9 |
|
GMC-3 |
CALR L367T*46 |
16.7 |
11.6 |
|
GMC-16 |
CALR K385Nfs*47 |
15 |
14.7 |
|
GMC-22 |
CALR K385Nfs*47 |
8.8 |
7.7 |
|
GMC-5 |
CSF3R T618I |
38.3 |
39.6 |
|
GMC-37 |
CSF3R T618I |
34.6 |
34.3 |
|
GMC-87 |
CSF3R T618I |
11.3 |
11.1 |
|
GMC-2 |
JAK2 V617F |
55.1 |
20.2 |
|
GMC-8 |
JAK2 V617F |
42.9 |
45.3 |
|
GMC-11 |
JAK2 V617F |
34.7 |
33.2 |
|
GMC-13 |
JAK2 V617F |
27.3 |
29.7 |
|
GMC-14 |
JAK2 V617F |
9.7 |
11.8 |
|
GMC-19 |
JAK2 V617F |
8.5 |
8 |
|
GMC-38 |
JAK2 V617F |
8.3 |
8.5 |
|
GMC-56 |
JAK2 V617F |
4.5 |
6.7 |
|
GMC-7 |
MPL R592* |
50.9 |
49.6 |
|
GMC-55 |
MPL W515K |
45.8 |
47.2 |
|
GMC-90 |
MPL S505N |
9.1 |
10.1 |
|
GMC-93 |
MPL S505N |
8.7 |
10.5 |
| Gene | Cases | VAF (%) |
|
JAK2 |
8/8 |
4.5-55.1 |
|
CALR |
4/4 |
7.7-51.9 |
|
MPL |
4/4 |
8.7-50.9 |
|
CSF3R |
3/3 |
11.3-38.3 |
|
IDH1 |
4/4 |
4.6-46.8 |
|
IDH2 |
5/3 |
34-46.2 |
|
SH2B3 |
1/1 |
47.2 |
|
NRAS |
4/5 |
4.4-45.8 |
|
TP53 |
3/3 |
8.4-75.9 |
Envisioning the Future of Molecular Diagnostics
Given their comparable sensitivity and their inherent advantages over single-gene testing in terms of efficiency, speed, and cost, Dr. Ding expects that NGS for multigene panel tests will continue to grow in the future, creating a new paradigm in molecular testing.
Learn More: Watch Dr. Ding’s AMP Workshop Presentation
Precision medicine is rapidly changing our understanding of cancer research and treatment decisions. These breakthrough, personalized treatments hold promise even for patients with historically hard-to-treat diseases, like lung or breast cancer. But
In his recent talk at OncomineWorld 2023, Dr. Bekim Sadikovic presented a strong, evidence-based argument for frontline next-generation sequencing (NGS) in myeloid malignancy testing.
Bekim Sadikovic PhD, DABMGG, FACMGProfessor, Research Chair, and...
In a recent Association for Molecular Pathology (AMP) workshop, Dr. Cecilia Yeung shared how her lab is implementing rapid next-generation sequencing (NGS) to help advance hemato-oncology research and expand access to underserved populations.
...