The theme of the 35th European Congress of Pathology (ECP) “Pathology – a bridge between science and medicine” was prominently evident throughout the renowned congress for the latest pathology updates in Europe.
This synergy between technology and medicine was particularly apparent in precision oncology, with research showcasing technological progress, novel approaches, and artificial intelligence applications for advancing various disciplines in the field of pathology.
One such advancement is the development of novel approaches for measuring homologous recombination deficiency (HRD). On the one hand, there is a need for HRD testing in research on ovarian and other cancer types; on the other hand, there is currently only one reference method, which is not broadly available.
Development of a novel metric to measure genomic instability using unbalanced copy number changes with fast comprehensive genomic profiling (PS-24-005)1. This is a poster presentation detailing a novel approach for measuring HRD, by estimating genomic instability based on unbalanced copy number changes.
Materials and methods
The Oncomine Comprehensive Assay Plus is a research use only (RUO) assay that detects DNA- and RNA-based pan-cancer biomarkers from over 500 genes, and complex biomarkers including tumor mutational burden (TMB), microsatellite instability (MSI), and HRD. In brief, 30 ng of DNA was used to prepare libraries with the Oncomine Comprehensive Assay Plus DNA panel. Library preparation is based on Ion AmpliSeq™ technology and sequencing was performed on the GeneStudio S5 System, or automated library preparation and sequencing was performed on the Genexus™ System (in development). Performance was characterized using control and formalin-fixed paraffin embedded (FFPE) samples.
Genomic Instability Metric and homologous recombination deficiency
Genomic instability metric (GIM) is a novel metric to quantify genomic scars/instability associated with HRD. It is based on genomic segmentation2 using CNV log2 ratios and log odds for SNP allele frequencies, which allows to summarize different unbalanced copy number events across autosomes. It ranges from 0-100, and higher values indicate more genomic instability.
Oncomine Comprehensive Assay Plus covers causes and consequences for HRD by interrogating 46 homologous recombination repair (HRR) pathway genes and estimating genomic instability through GIM. BRCA1 and BRCA2 are enabled for SNV/indel calling as well as CNV, gene LOH and large genomic rearrangements such as exon-level deletions and duplications.
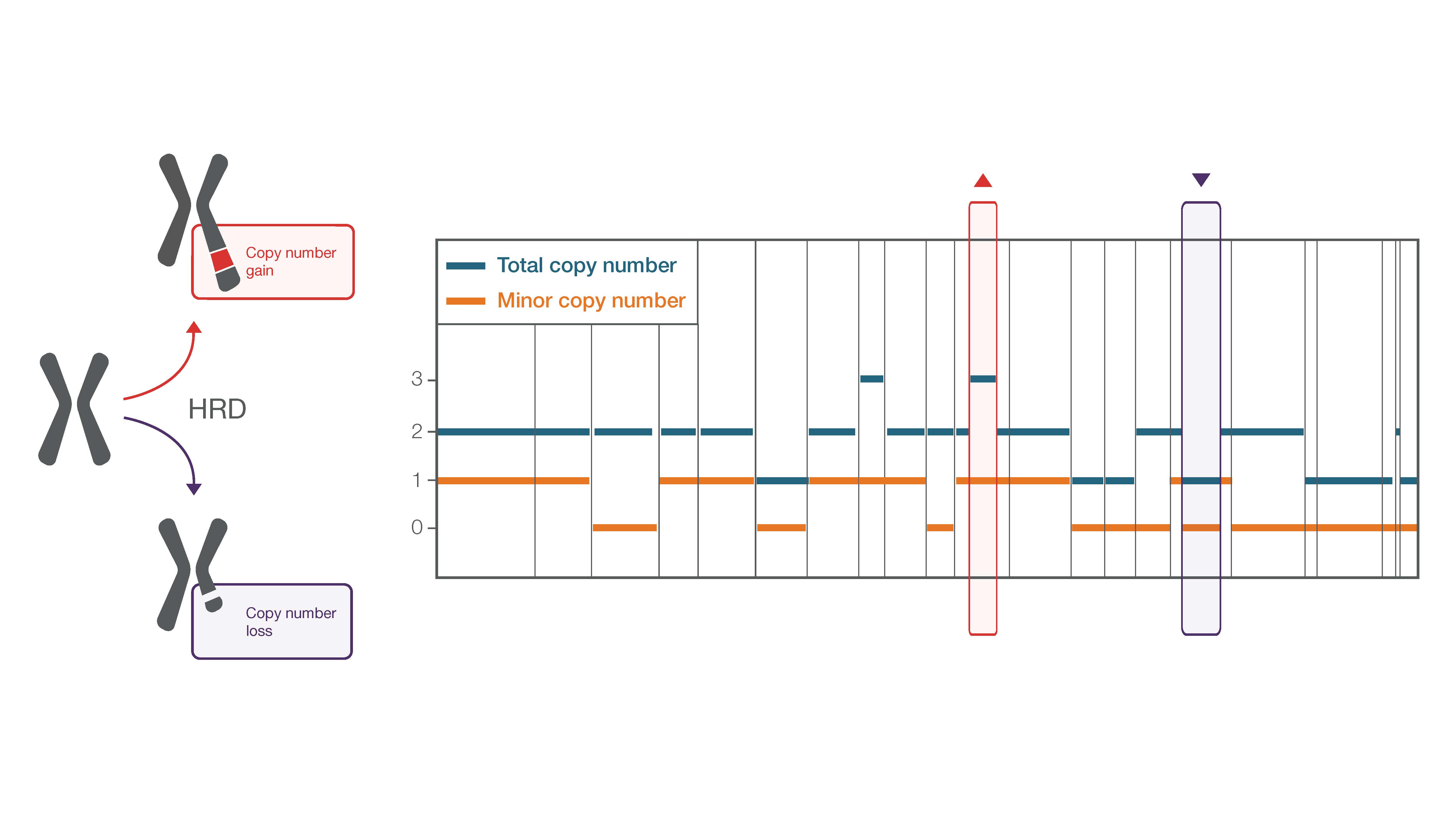 Figure 1. Schematic example depicting how a copy number gain and loss are affecting minor and total copy numbers.
Figure 1. Schematic example depicting how a copy number gain and loss are affecting minor and total copy numbers.
Results
Ovarian cancer samples with pathogenic BRCA1/2 mutations are HRD positive. GIM can stratify BRCA1/2 mutated samples from BRCA1/2 wild-type samples with a threshold of 16 for determining HRD status in ovarian cancer samples.
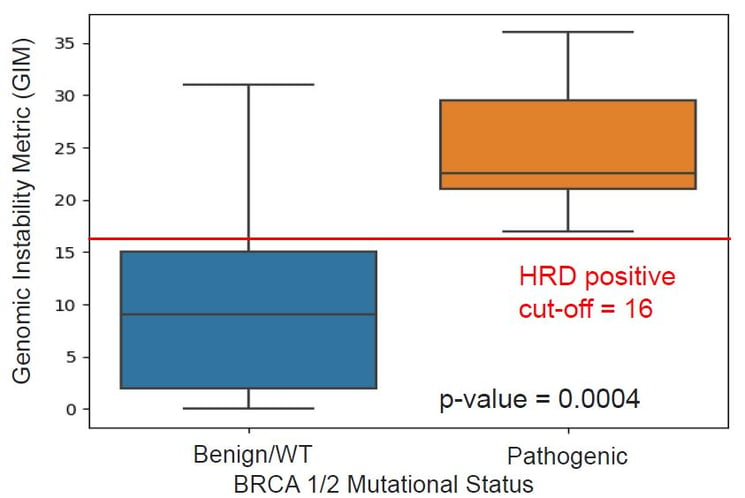
Figure 2. Stratification of HRD+ samples by GIM
85 ovarian FFPE samples were sequenced on the GeneStudio S5. Results from the Oncomine Comprehensive Assay Plus (GIM and BRCA1/2 mutational status) was compared to a reference assay and summarized in the Table below. The study demonstrated an agreement of 90.7%.3
| OCA Plus HRD+ | OCA Plus HRD- | |
| Reference HRD+ | 51 | 1 |
| Reference HRD- | 7 | 27 |
| Sensitivity | 98.1% |
| Specificity | 79.4% |
| Agreement | 90.7% |
Conclusion:
Access to HRD testing in many countries remains a challenge due to cost, lengthy turnaround times, and/or high failure rates. The Oncomine Comprehensive Assay Plus is a targeted, next-generation sequencing assay that provides comprehensive genomic profiling for FFPE tissues from solid tumors, including small and degraded samples.
With the development of GIM, HRD status could be assessed simultaneously with mutations in HRR pathway genes without requiring any additional wet or dry lab effort aside from the Oncomine Comprehensive Assay Plus workflow.
For the full poster and additional activities from ECP 2023, please visit https://www.oncomine.com/ecp2023.
References:
1. M. Gupta et al. Poster PS-24-005, ECP 2023.
2. S. Ronglai et al., 2016, Nucleic Acids Research, Vol.44, No.16
3. C. Roma et al., Poster abstract EACR23-1096, EACR Congress 2023.
For Research Use Only. Not for use in diagnostics procedures.
November 3, 2023, 49275
In everyday practice, it is common for pathology labs to evaluate >50% cytological specimens across solid tumors especially in lung and pancreatic cancers. Many of these cytological specimens require molecular profiling to help confirm diagnoses and...
The theme of the 35th European Congress of Pathology (ECP) “Pathology – a bridge between science and medicine” was prominently evident throughout the renowned congress for the latest pathology updates in Europe.
This synergy between technology and...
In a recent Association for Molecular Pathology (AMP) workshop, Dr. Cecilia Yeung shared how her lab is implementing rapid next-generation sequencing (NGS) to help advance hemato-oncology research and expand access to underserved populations.
...