In recent years, we’ve seen an increasing number of new therapies emerge that target gene fusions in a range of cancer types, and there’s more in the pharmaceutical pipeline every day. A great example is the drug larotrectinib (Loxo Oncology, Bayer), which fights a range of solid tumor types by attacking TRK—a protein that’s hyperactive in people with certain NTRK gene fusions.1 The efficacy of larotrectinib has been remarkable—with an 80% objective response rate (ORR) across multiple tumor types.2 This statistic alone illustrates why it’s so critical to test oncology patients for gene fusions and markers of targeted therapies.
As a result, many labs are considering options for fusion detection to ensure they’re well equipped to profile these key biomarkers. In this article, I’m going to review the fundamentals of gene fusions and discuss various methods used to detect them. I’ll also provide some tips on things to consider before you decide on the best method for your lab.
Gene fusions and cancer
Let’s start with the basics. Gene fusions occur when 2 genes that are normally not connected fuse at a specific break point, resulting from chromosomal translocations, rearrangements, or deletions.3–5 The driver gene initiates the fusion with the partner gene (Figure 1). Any driver gene can contain numerous different partners and breaking points known as isoforms. Similar to mutations, fusions are another variant type associated with cancers. Clinically relevant gene fusions were originally discovered in patients with chronic myelogenous leukemia.5–7 Subsequently, one of the first precision medicines in cancer treatment for chronic myelogenous leukemia (CML) was approved in 2001.8 Over the last 20 years, many gene fusions have been discovered in hematological cancers, solid tumors, and sarcomas, and have become important biomarkers for cancer diagnosis, prognosis, and targeted therapies.1,9,10
The most common example of a gene fusion in solid tissue cancer is EML4-ALK. The discovery of the fusion between echinoderm microtubule-associated protein-like 4 (EML4) and anaplastic lymphoma kinase (ALK), and the subsequent identification of at least 15 different variants in lung cancers were a huge leap forward for precision oncology.2 For instance, molecular screening is now a routine procedure for nonsmall cell lung cancer (NSCLC), which comprises more than 80% of all lung cancers.2
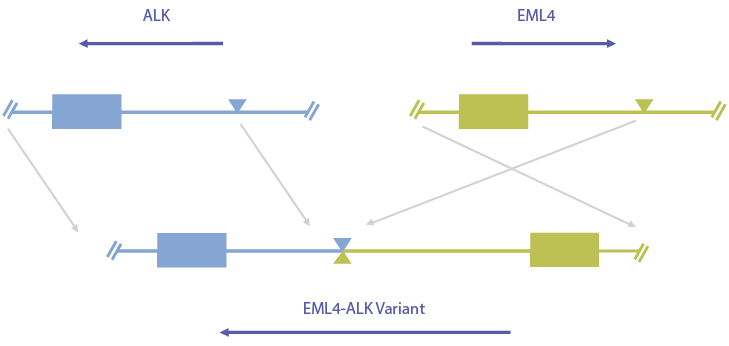
Figure 1. The driver gene initiates the event, usually from the 3-prime end of the gene—ALK in this case.
Detection methods
Supporting this type of clinical research requires a robust and reliable detection technology. There are multiple methods to test for fusions, from traditional single-gene testing to the more-advanced and more-efficient multimarker, next-generation sequencing (NGS).
Among them are fluorescence in situ hybridization (FISH), immunohistochemistry (IHC), and reverse transcriptase-polymerase chain reaction (RT-PCR). The major disadvantage of these is that they’re limited to single-gene (or single-isoform in the case of RT-PCR) tests. Traditional single-gene testing, as the name indicates, entails testing 1 marker at a time. Single-gene tests take longer and may completely diminish the limited sample without generating a useful result. With the growing number of relevant biomarkers, sequential single-gene testing becomes increasingly impractical, particularly when you consider time and tissue limitations.
NGS
NTRK is a perfect example of a low-prevalence fusion that might go untested with traditional methods, as tissue is likely to be used in testing for more prevalent markers. NGS, on the other hand, can be used in clinical laboratories for simultaneous detection of diverse somatic variants in cancer, including gene fusions.11,12 Hands-down (literally, with the new automated workflows), NGS is the most practical, efficient, and effective technology for analyzing multiple biomarkers at the same time. It enables you to look at both common and rare fusions from either a tissue or blood sample. When it comes to precision oncology, the choice between traditional single-gene testing and multiple-gene testing with NGS is clear.
NGS key considerations
It’s important to recognize that not all NGS is created equal. When it comes to maximizing fusion detection, the NGS goals should be to test as many samples as possible, achieve sensitive and specific performance for the most-prevalent fusion isoforms, and detect novel fusion isoforms.
Test as many samples as possible
The first critical NGS consideration is the sample. Unfortunately, many NGS technologies require both greater nucleic acid inputs and/or higher tumor content, which immediately excludes a large portion of samples. This results in a significant percentage of samples that are either rejected before testing starts or receive a quantity not sufficient (QNS) result due to lack of sufficient tissue or tumor. These combined factors dramatically decrease the overall number of samples successfully tested. Some would say, unacceptably. That’s why it’s so critical to seek out NGS with low sample input requirements, which means you’ll be able to test more of those precious samples out of the gate. When it comes to maximizing the number of fusions you can detect, low sample input is essential.
Sensitive and specific performance for the most-prevalent fusion isoforms
The other key factor for maximizing fusion detection is high sensitivity and specificity—the ability to detect low-frequency variants without introducing false positives—for the most-prevalent fusion isoforms. For many fusion driver genes, a handful of known isoforms make up a majority of fusions that are detected (eg, ETV6-NTRK3, EML4-ALK, KIF5B-RET). For these prevalent isoforms, having targeted designs (with known driver, partner, and breakpoint) is preferable, as analytical pipelines can be tuned to maximize performance, especially since these fusion isoforms don’t exist in normal background.
Capability of detecting novel fusion isoforms
While a majority of fusions are known and prevalent isoforms, there’s also the presence of novel fusion isoforms, or those that have yet to be discovered and documented in public literature. This is especially true for certain driver genes that tend to be more “promiscuous.” Novel isoforms can exist in the form of new combinations of drivers and partners, or completely unknown partners and breakpoints of an existing driver gene. While having the capability to detect these more-infrequent novel isoforms is important, it can’t be at the expense of the other key considerations mentioned above.
Bottom line
The primary goal remains to test as many samples as possible for known fusions. While rare and novel fusions can provide useful information, they shouldn’t be prioritized at the expense of testing fewer samples. As NGS for fusion detection becomes ubiquitous, it will be possible to rapidly advance the promise of precision oncology.
REFERENCES
1. Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846.
2. Sabir SR, Yeoh S, Jackson G, Bayliss R. EML4-ALK variants: biological and molecular properties, and the implications for patients. Cancers (Basel). 2017;9(9):118.
3. Parker BC, Zhang W. Fusion genes in solid tumors: an emerging target for cancer diagnosis and treatment. Chin J Cancer. 2013;32(11):594-603.
4. Gao Q, Liang WW, Foltz SM, et al. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep. 2018;23(1):227-238.
5. Kumar-Sinha C, Kalyana-Sundaram S, Chinnaiyan AM. Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome Med. 2015;7:129.
6. Koretzky GA. The legacy of the Philadelphia chromosome. J Clin Invest. 2007;117(8):2030-2032.
7. Stam K, Heisterkamp N, Grosveld G, et al. Evidence of a new chimeric bcr/c-abl mRNA in patients with chronic myelocytic leukemia and the Philadelphia chromosome. N Engl J Med. 1985;313(23):1429-1433.
8. Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112(13):4808-4817.
9. Schram AM, Chang MT, Jonsson P, Drilon A. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat Rev Clin Oncol. 2017;14(12):735-748.
10. Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15(6):371-381.
11. Aziz N, Zhao Q, Bry L, et al. College of American Pathologists' laboratory standards for next-generation sequencing clinical tests. Arch Pathol Lab Med. 2015;139(4):481-493.
12. Jennings LJ, Arcila ME, Corless C, et al. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017;19(3):341-365.

David Chi is a Sr. Product Manager in the clinical NGS and oncology division at Thermo Fisher Scientific. Prior to this role, he was a program manager supporting both product development and clinical laboratory operations. Before Thermo Fisher Scientific, he had a wide range of experience in the medical device field.
A recently published article by the BLOODPAC Consortium details the potential of liquid biopsy in the management of cancer and highlights the barriers to adoption, particularly in underserved populations.
Cancer is one of the leading causes of...
Precision medicine is rapidly changing our understanding of cancer research and treatment decisions. These breakthrough, personalized treatments hold promise even for patients with historically hard-to-treat diseases, like lung or breast cancer. But
Dr. Eric Vail is Director of Molecular Pathology, Cedars-Sinai Medical Center,Los Angeles, California
Can you please introduce yourself and your laboratory?
I'm the director of the Cedar Sinai Medical Center Molecular Pathology Laboratory, which...