Structural variants in the cancer genome
Gene amplification (copy number gain) and deletion (copy number loss) are common in cancer cells and contribute to cancer cell growth, drug sensitivity and resistance. Herceptin (trastuzumab), one of the first targeted precision medicines for cancer, was designed to treat breast cancer patients whose tumors contain ERBB2 (HER2) gene amplification.
Genes like ERBB2 (HER2) that promote cancer progression (oncogenes) often experience an abnormal increase in their gene count. In contrast, genes that restrain cancer progression (tumor suppressor genes) may be removed from the genome by gene deletion.
The gain or loss of gene copies often correlates with a corresponding increase or decrease in the amount of RNA and protein encoded by the gene. Being able to efficiently assess copy number changes in tumor samples is important to support both translational research and foster precision medicine.
Development of methods to detect gene amplification and deletion
The discovery of gene amplification in mammalian cells is attributed to Schimke and colleagues who observed that an important mechanism of resistance to the anti-cancer drug methotrexate involved the amplification of the gene encoding dihydrofolate reductase (1, 2, 3).
Researchers soon after discovered that genes subjected to amplification in cancer were often the proto-oncogenes that had cellular homologs resident in the genomes of certain transforming viruses (4,5,6). In parallel, researchers discovered that a different set of genes were frequently lost from the genome of certain cancers (7, 8, 9).
Subsequent research revealed that gene amplification and deletion were widespread and prevalent in the human cancer genome (10, 11, 12). Over the years, several methods were used to investigate changes in gene copy number including cytogenetics, southern blot hybridization, fluorescence in-situ hybridization (FISH), polymerase chain reaction (PCR), comparative genomic hybridization (CGH) and whole genome single-nucleotide polymorphism (SNP) arrays (Figure 1). Some of these methods such as PCR and FISH are fast, but not readily scalable to many targets.
Other methods, including arrays, are scalable to many targets but require large sample volumes to provide sufficient DNA. Recently, comprehensive genomic profiling using next-generation sequencing (NGS) has extended research into somatic copy number alterations (13, 14). Relative to other methods, NGS has the advantage of providing a rapid solution for multiple targets using limited sample input.
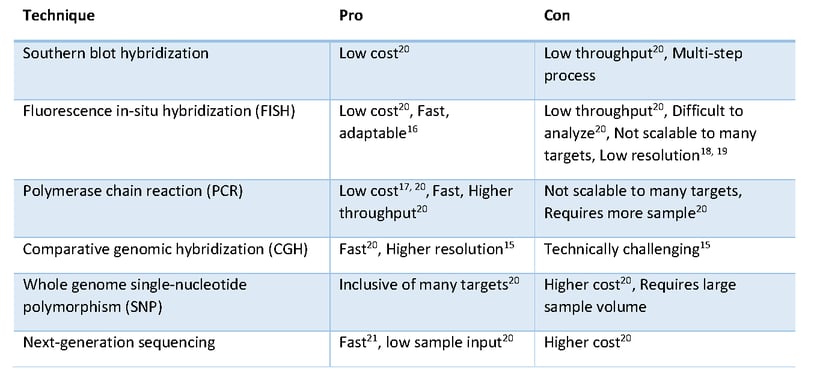
Figure 1: CNV Detection Techniques
The landscape of copy number variation in cancer
Copy number variation encompasses a wide range of structural alterations in the genome (Figure 2). In cancer, it includes focal changes in localized genomic regions involving a small number of genes as well as large alterations such as the gain or loss of an entire chromosomal arm and having an abnormal number of chromosomes in a cell.
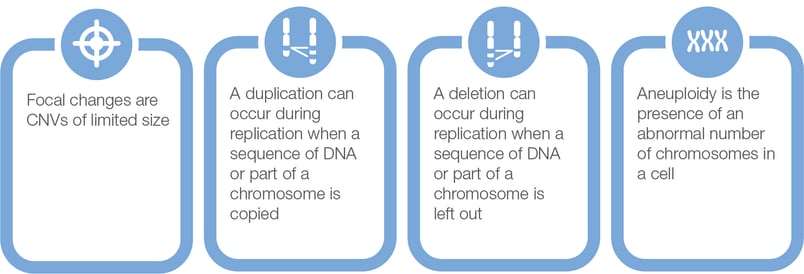
Figure 2: CNV Types
Changes in chromosome number and chromosome arms are collectively referred to as aneuploidy. Focal copy number changes and aneuploidy are common in cancer and carry important information for clinical research, which can in future be related to diagnosis, prognosis, and therapeutic response. For example, loss of chromosomal arms 1p and 19q are closely associated with oligodendrogliomas (22), a subtype of primary brain tumors, and with favorable prognosis in diffuse gliomas. Focal copy number changes may be predictive biomarkers for certain therapeutics (for example, ERBB2 amplification in breast cancer) and can be selected for as part of a mechanism of acquired resistance (for example, MET amplification in NSCLC) (23, 24, 25, 26).
A survey of focal copy number alterations in different cancer types shows that certain genes are commonly amplified or deleted in several cancer types whereas other genes tend to be altered in specific cancer types (see link to Table 1 at the end of the blog).
Using NGS to estimate copy number variation in cancer research samples
Relative to other somatic alterations, copy number variation is relatively straightforward to detect using NGS. The gain or loss of gene copies can be reliably detected by the gain or loss of sequencing reads corresponding to genomic regions of interest. Hence, copy number variation using NGS is essentially a read-counting application.
In Ion Torrent Oncomine targeted amplicon sequencing, the first step is to use Ion AmpliSeq PCR technology to enable the multiplexing of thousands of PCR reactions for the uniform amplification of DNA targets. The PCR reaction is then subjected to barcoding, templating, and sequencing. Sequence reads are aligned to the genomic reference and counted. In some cases of high-level gene amplification, the accumulation of sequence reads to specific targets can be observed in the raw data.
However, because each targeted amplicon has its own characteristic amplification efficiency, normalization strategies are used to address intrinsic (amplicon length, GC content) and extrinsic (sample and assay effects) sources of variability. To address sample and assay effects, a principal components approach is used to characterize variability using an extensive training set of libraries prepared from a diverse set of samples that is saved as a pre-computed informatics baseline.
Each Oncomine NGS workflow includes a baseline to automatically correct and normalize read counts and deliver improved copy number (CN) estimates for every gene. Occasionally, samples are identified with high levels of variability that preclude CN estimation. In these cases, typically due to low sample quality, a metric “median absolute pairwise difference” (MAPD) is useful to identify samples that are at risk of loss of sensitivity or generating false results.
Hybridization capture technology, an alternative NGS technology, can also be used for targeted sequencing with sensitivity down to 1% without unique molecular identifiers. As these panels can contain more targets and grow to almost unlimited size, they are suited for exome sequencing and genotyping in basic and translational research. However, as this technology, in order to achieve such high coverage, requires more sample, steps and time depending on the panel design, it might not be as applicable for routine clinical research.
An example of using Oncomine NGS to detect gene amplification in formalin-fixed paraffin embedded (FFPE) solid tumor samples is shown in Figure 3. A cancer research sample of glioblastoma was processed using the Ion Torrent Oncomine Focus Assay. The scatterplot shows the read counts associated with amplicons arranged on the x-axis in the order of their genomic location. In this glioblastoma sample, you can already see evidence of EGFR copy number gain even in the raw read counts. In the bottom panel, the corrected read counts are converted to log2 copy number ratios and you can see the data becomes much tighter. On the right are corresponding FISH images from sections of the same tumor block, showing high copy number gain for EGFR and no copy number gain for another gene, ERBB2 (HER2) in this case.
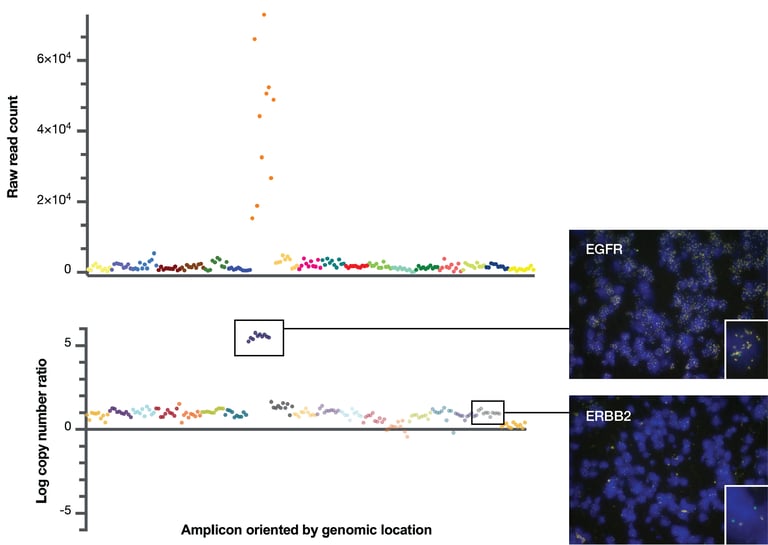
Figure 3: CNV estimation using Ion AmpliSeq and Ion Reporter
A summary of the normalized copy number data estimated in 20 different tumor samples and its concordance to FISH data is shown in Figure 4. In this experiment, we chose 20 FFPE samples that had copy number gains as predicted by NGS and then developed FISH assays against 10 different genes. We then estimated copy number by FISH in one lab and by NGS in the 4 different laboratories and compared the results.
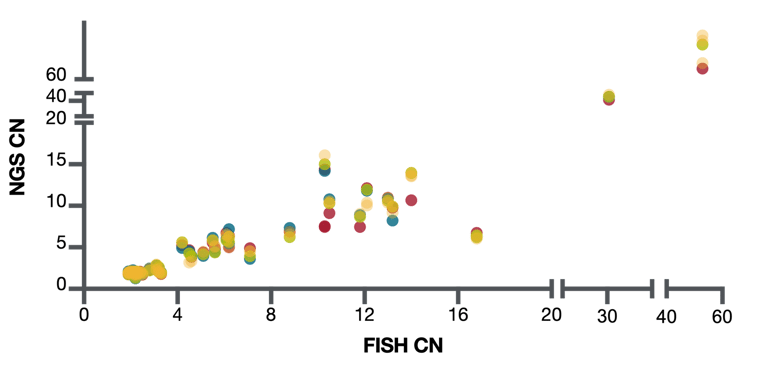
Figure 4: CNV detection NGS comparison to FISH
Overall, the concordance between copy number estimates as determined by FISH and by NGS was very good. There was also good reproducibility between the 4 different laboratories. From this experiment, we estimated our sensitivity to detect copy number gains of 8 and more at 97% and for detecting copy number gains of 6 and more at 94%. Importantly, there were no false positives in this experiment.
Conclusion
Identifying copy number alterations in tumor samples through NGS and other techniques supports precision oncology clinical research as copy number variants can be utilized for the diagnosis, prognosis and treatment of various cancer types. Oncomine NGS solutions support research aiming to better uncover the role of copy number changes in cancer. Oncomine NGS solutions assess CNVs well in comparison to other technologies such as FISH and have distinct advantages such as scalable targets, high throughput, and the ability to deliver a comprehensive genomic profile when analyzed with all other classes of genomic alterations.
References:
1: Schimke RT, Kaufman RJ, Alt FW, Kellems RF. Gene amplification and drug resistance in cultured murine cells. Science. 1978 Dec 8;202(4372):1051-5. doi: 10.1126/science.715457. PMID: 715457.
2: Schimke RT, Kaufman RJ, Nunberg JH, Dana SL. Studies on the amplification of dihydrofolate reductase genes in methotrexate-resistant cultured mouse cells. Cold Spring Harb Symp Quant Biol. 1979;43 Pt 2:1297-303. doi: 10.1101/sqb.1979.043.01.148. PMID: 290443.
3: Schimke RT, Brown PC, Kaufman RJ. Gene amplification and drug resistance in mammalian cells. Natl Cancer Inst Monogr. 1982;60:79-86. PMID: 7121575.
4: Collins S, Groudine M. Amplification of endogenous myc-related DNA sequences in a human myeloid leukaemia cell line. Nature. 1982 Aug 12;298(5875):679-81. doi: 10.1038/298679a0. PMID: 6285209.
5: Schwab M, Alitalo K, Klempnauer KH, Varmus HE, Bishop JM, Gilbert F, Brodeur G, Goldstein M, Trent J. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983 Sep 15-21;305(5931):245-8. doi: 10.1038/305245a0. PMID: 6888561.
6: Semba K, Kamata N, Toyoshima K, Yamamoto T. A v-erbB-related protooncogene, c-erbB-2, is distinct from the c-erbB-1/epidermal growth factor-receptor gene and is amplified in a human salivary gland adenocarcinoma. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6497-501. doi: 10.1073/pnas.82.19.6497. PMID: 2995967; PMCID: PMC390744.
7: Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997 Mar 28;275(5308):1943-7. doi: 10.1126/science.275.5308.1943. PMID: 9072974.
8: Teng DH, Hu R, Lin H, Davis T, Iliev D, Frye C, Swedlund B, Hansen KL, Vinson VL, Gumpper KL, Ellis L, El-Naggar A, Frazier M, Jasser S, Langford LA, Lee J, Mills GB, Pershouse MA, Pollack RE, Tornos C, Troncoso P, Yung WK, Fujii G, Berson A, Steck PA, et al. MMAC1/PTEN mutations in primary tumor specimens and tumor cell lines. Cancer Res. 1997 Dec 1;57(23):5221-5. PMID: 9393738.
9: Kleihues P, Lübbe J, Watanabe K, von Ammon K, Ohgaki H. Genetic alterations associated with glioma progression. Verh Dtsch Ges Pathol. 1994;78:43-7. PMID: 7534015.
10: Bishop JM. The molecular genetics of cancer. Science. 1987 Jan 16;235(4786):305-11. doi: 10.1126/science.3541204. PMID: 3541204.
11: Kashiwagi H, Uchida K. Genome-wide profiling of gene amplification and deletion in cancer. Hum Cell. 2000 Sep;13(3):135-41. PMID: 11197775.
12: Brodeur GM. The involvement of oncogenes and suppressor genes in human neoplasia. Adv Pediatr. 1987;34:1-44. PMID: 3318293.
13: Singh RR, Patel KP, Routbort MJ, Aldape K, Lu X, Manekia J, Abraham R, Reddy NG, Barkoh BA, Veliyathu J, Medeiros LJ, Luthra R. Clinical massively parallel next-generation sequencing analysis of 409 cancer-related genes for mutations and copy number variations in solid tumours. Br J Cancer. 2014 Nov 11;111(10):2014-23. doi: 10.1038/bjc.2014.518. Epub 2014 Oct 14. PMID: 25314059; PMCID: PMC4229640.
14: Grasso C, Butler T, Rhodes K, Quist M, Neff TL, Moore S, Tomlins SA, Reinig E, Beadling C, Andersen M, Corless CL. Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J Mol Diagn. 2015 Jan;17(1):53-63. doi: 10.1016/j.jmoldx.2014.09.008. Epub 2014 Nov 7. PMID: 25468433; PMCID: PMC5707205.
1 15: Redon R, Carter NP. Comparative genomic hybridization: microarray design and data interpretation. Methods Mol Biol. 2009;529:37-49. doi:10.1007/978-1-59745-538-1_3
16: Volpi EV, Bridger JM. FISH glossary: an overview of the fluorescence in situ hybridization technique. Biotechniques. 2008 Oct;45(4):385-6, 388, 390 passim. doi: 10.2144/000112811.
17: NIH, National Human Genome Research Institute, Polymerase Chain Reaction (PCR) Fact Sheet, Updated: August 17, 2020
18: Zhang L, Bai W, Yuan N, Du Z. Correction: Comprehensively benchmarking applications for detecting copy number variation. PLoS Comput Biol. 2019;15(9):e1007367. Published 2019 Sep 20. doi:10.1371/journal.pcbi.1007367
19: Duan J, Zhang JG, Deng HW, Wang YP. Comparative studies of copy number variation detection methods for next-generation sequencing technologies. PLoS One. 2013;8(3):e59128. doi:10.1371/journal.pone.0059128
20: Cantsilieris S, Baird PN, White SJ. Molecular methods for genotyping complex copy number polymorphisms. Genomics. 2013 Feb;101(2):86-93. doi: 10.1016/j.ygeno.2012.10.004. Epub 2012 Oct 30. PMID: 23123317.
21: https://www.thermofisher.com/genexus
22: McDonald JM, See SJ, Tremont IW, Colman H, Gilbert MR, Groves M, Burger PC, Louis DN, Giannini C, Fuller G, Passe S, Blair H, Jenkins RB, Yang H, Ledoux A, Aaron J, Tipnis U, Zhang W, Hess K, Aldape K. The prognostic impact of histology and 1p/19q status in anaplastic oligodendroglial tumors. Cancer. 2005 Oct 1;104(7):1468-77. doi: 10.1002/cncr.21338. PMID: 16088966.
23: Pegram MD, Pauletti G, Slamon DJ. HER-2/neu as a predictive marker of response to breast cancer therapy. Breast Cancer Res Treat. 1998;52(1-3):65-77. doi: 10.1023/a:1006111117877. PMID: 10066073.
24: Pegram MD, Konecny G, Slamon DJ. The molecular and cellular biology of HER2/neu gene amplification/overexpression and the clinical development of herceptin (trastuzumab) therapy for breast cancer. Cancer Treat Res. 2000;103:57-75. doi: 10.1007/978-1-4757-3147-7_4. PMID: 10948442.
25: Chung CT, Yeh KC, Lee CH, Chen YY, Ho PJ, Chang KY, Chen CH, Lai YK, Chen CT. Molecular Profiling of Afatinib-Resistant Non-Small Cell Lung Cancer Cells in vivo Derived from Mice. Pharmacol Res. 2020 Sep 4:105183. doi: 10.1016/j.phrs.2020.105183. Epub ahead of print. PMID: 32896579.
26: Nishiyama A, Takeuchi S, Adachi Y, Otani S, Tanimoto A, Sasaki M, Matsumoto S, Goto K, Yano S. MET amplification results in heterogeneous responses to osimertinib in EGFR-mutant lung cancer treated with erlotinib. Cancer Sci. 2020 Jul 31. doi: 10.1111/cas.14593. Epub ahead of print. PMID: 32735723.
A recently published article by the BLOODPAC Consortium details the potential of liquid biopsy in the management of cancer and highlights the barriers to adoption, particularly in underserved populations.
Cancer is one of the leading causes of...
Precision medicine is rapidly changing our understanding of cancer research and treatment decisions. These breakthrough, personalized treatments hold promise even for patients with historically hard-to-treat diseases, like lung or breast cancer. But
Dr. Eric Vail is Director of Molecular Pathology, Cedars-Sinai Medical Center,Los Angeles, California
Can you please introduce yourself and your laboratory?
I'm the director of the Cedar Sinai Medical Center Molecular Pathology Laboratory, which...